Extremophiles are organisms that can thrive in extreme environmental conditions of various physical and chemical parameters such as temperature, pressure, pH, and salinity. Among the extremophiles, psychrophiles and thermophiles, which are tolerant of low and high temperatures, respectively, have been of particular interest as they have potential in biotechnological applications. Proteins isolated from psychrophilic and thermophilic organisms can remain physiologically functional at extreme temperatures, which are detrimental to mesophilic proteins from organisms living at moderate temperatures. Hence, psychrophilic and thermophilic proteins are desirable for use in many academic and industrial settings that require biological activity at the extreme temperatures.
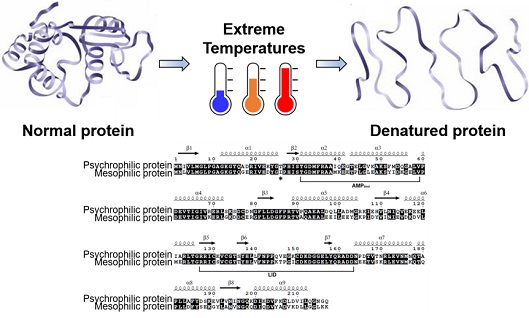
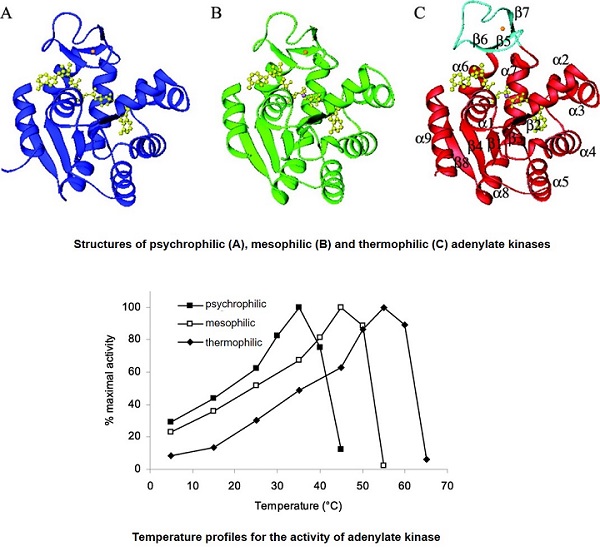
To evaluate the molecular basis for the temperature adaptation of psychrophilic and thermophilic proteins, we have performed comparisons with their mesophilic homologues. Using adenylate kinases as models, we reported crystal structures, thermal denaturation midpoints, temperature-activity profiles, and molecular dynamics trajectories of protein homologues from psychrophilic, mesophilic, and thermophilic organisms. Modifications of intramolecular interactions such as hydrogen bonds, electrostatic interactions, and hydrophobic contacts have been identified as the main structural mechanisms of cold and heat adaptation, but the various individual structural features change unpredictably to different extents in different proteins. The alterations of intramolecular interactions are the results of structural adjustments that allow for appropriate flexibility to ensure physiological function at different temperatures.
Related publications:
1. Kim, J., Moon, S., Romo, T.D., Yang, Y., Bae, E.*, Phillips, G.N.* (2024) "Conformational dynamics of adenylate kinase in crystals." Struct Dyn 11(1): 014702.
2. Moon, S.#, Kim, J.#, Koo, J. and Bae, E.* (2019) “Structural and mutational analyses of psychrophilic and mesophilic adenylate kinases highlight the role of hydrophobic interactions in protein thermal stability.” Struct Dyn 6(2): 024702.
3. Moon, S.#, Kim, J.# and Bae, E.* (2017) “Structural analyses of adenylate kinases from Antarctic and tropical fishes for understanding cold adaptation of enzymes.” Sci Rep 7(1): 16027.
4. Bae, E.*, Moon, S. and Phillips G.N., Jr. (2015) “Molecular dynamics simulation of a psychrophilic adenylate kinase.” J Korean Soc App Biol Chem 58(2): 209-212.
5. Moon, S. and Bae, E.* (2014) “Crystal structures of thermally stable adenylate kinase mutants designed by local structural entropy optimization and structure-guided mutagenesis.” J Korean Soc App Biol Chem 57(5): 661-665.
6. Moon, S., Bannen. R.M., Rutkoski, T.J., Phillips G.N., Jr. and Bae, E.* (2014) “Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.” Proteins 82(10): 2631-2642.
7. Moon, S., Jung, D., Phillips, G.N., Jr. and Bae, E.* (2014) “An integrated approach for thermal stabilization of a mesophilic adenylate kinase.” Proteins 82(9): 1947-1959.