Clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins provide adaptive immunity in bacteria and archaea for defense against invading foreign nucleic acids such as bacteriophages and plasmids. To counteract the microbial CRISPR-mediated immunity, phages have evolved anti-CRISPR (Acr) proteins that inhibit the host CRISPR-Cas systems.
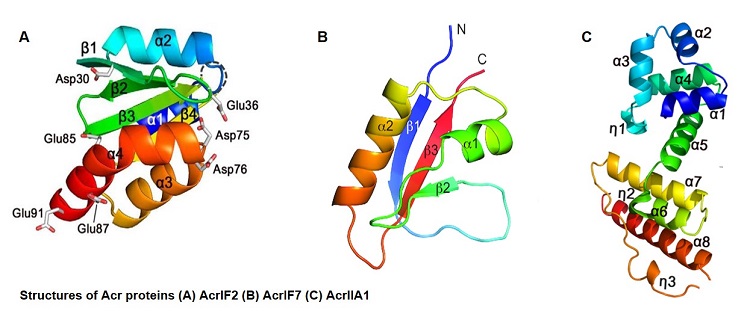
We have reported structure, function and interaction of various type I and type II Cas proteins. We have also investigated several Acr proteins such as AcrIF2, AcrIIA1 and AcrIF7. Structural and biochemical analyses of the Acr proteins revealed diverse inhibitory mechanisms between Acr proteins. Understanding the structural and inhibitory mechanisms of novel Acr proteins would provide a better opportunity to control CRISPR-Cas function for biotechnological applications such as genome editing.
Related publications:
1. Koo, J., Lee, G., Ka, D., Park, C., Suh, J.Y., Bae, E.* (2023) "Biochemical characterization of type I-E anti-CRISPR proteins, AcrIE2 and AcrIE4." Appl Biol Chem 66, 51.
2. Hong, S.H.#, Lee, G.#, Park, C., Koo, J., Kim, E.H., Bae, E.*, Suh, J.Y.* (2022) “The structure of AcrIE4-F7 reveals a common strategy for dual CRISPR inhibition by targeting PAM recognition sites.” Nucleic Acids Res 50(4):2363-2376.
3. Kim, I.#, Koo, J.#, An, S.Y.#, Hong, S., Ka, D., Kim, E.H., Bae, E.* and Suh, J.Y.* (2020) “Structural and mechanistic insights into the CRISPR inhibition of AcrIF7.” Nucleic Acids Res 48(17): 9959-9968.
4. Ka, D., Jang, D.M., Han, B.W. and Bae, E.* (2018) “Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2.” Nucleic Acids Res 46(18): 9805-9815.
5. Hong, S.#, Ka, D.#, Yoon, S.J., Suh, N., Jeong, M., Suh, J.Y. and Bae, E.* (2018) “CRISPR RNA and anti-CRISPR protein binding to the Xanthomonas albilineans Csy1-Csy2 heterodimer in the type I-F CRISPR-Cas system.” J Biol Chem 293(8): 2744-2754.
6. Ka, D.#, An, S.Y.#, Suh, J.Y.* and Bae, E.* (2018) “Crystal structure of an anti-CRISPR protein, AcrIIA1.” Nucleic Acids Res 46(1): 485-492.
7. Ka, D., Hong, S., Jeong, U., Jeong, M., Suh, N., Suh, J.Y. and Bae, E.* (2017) “Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity.” Struct Dyn 4(5): 054701.
8. Ka, D., Lee, H., Jung, Y.D., Kim, K., Seok, C., Suh, N. and Bae, E.* (2016) “Crystal Structure of Streptococcus pyogenes Cas1 and Its Interaction with Csn2 in the Type II CRISPR-Cas System.” Structure 24(1): 70-79.
9. Ka, D., Kim. D., Baek, G. and Bae, E.* (2014) “Crystal Structure of Streptococcus pyogenes Cas1 and Its Interaction with Csn2 in the Type II CRISPR-Cas System. Biochem Biophys Res Commun 451(1):152-157.
10. Yoon, K.#, Ka, D.#, Kim, E.J., Suh, N.* and Bae, E.* (2013) "Conservation and variability in the structure and function of the Cas5d endoribonuclease in the CRISPR-mediated microbial immune system.” J Mol Biol 425(20): 3799-3810.
11. Yoon, K.#, Jung, D.# and Bae, E.* (2012) "Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure." PLoS One 7(3): e33401.